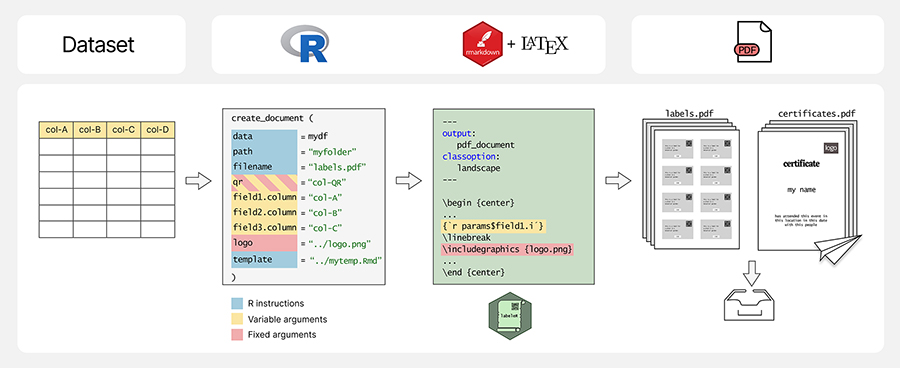
Figure 2. Examples of the outcomes from each label-related function in
labeleR. a) Herbarium label: for stored plant vouchers; includes fixed fields
(title, subtitle) and variable fields (e.g. taxon, date, coordinates,
elevation). The family field is by default capitalized and bold, and species
name italic. Size: 4 labels/page. b) Collection label: includes variable fields
(first field in italics, second in bold), a customizable logo, font and
background and text colors. Size: 8/page. c) Tinylabel: a simplified collection
label with five fields. Size: 16/page. All three functions can include an
optional QR code.
Figura
2. Ejemplos de salida de las funciones relacionadas con etiquetas en
labeleR. a) etiquetas de herbario: para pliegos de herbario se incluyen campos
fijos (título y subtítulo) y campos variables por cada especímen
(e.g. taxón, fecha, coordenadas, elevación). El campo de la familia es por
defecto en mayúsculas y negrita, mientras que la especie aparece en cursiva.
Tamaño: 4 etiquetas/página. b) Etiqueta de colección: incluye campos variables
(el primero en cursiva, el segundo en negrita), y otros modificables como el
logo, la fuente y los colores de fondo y de texto. Tamaño: 8/página. c)
Tinylabel: es una etiqueta de colección simplificada con solo cinco campos.
Tamaño: 16/página. En las tres funciones se puede incluir un QR.
Documents for scientific
events
Scientific
events often host a high number of participants, and require the creation of
identification badges, abstract books and certificates for attendees and
participants (Fig. 3). With labeleR, templates and code
can be customized according to the event, and bulk rendering significantly
decreases the amount of time invested in the creation of such materials.
Manually delivering attendance and participation certificates is highly
time-consuming, but labeleR functions allow
users to automatically send individual documents to email addresses stored in a
column.
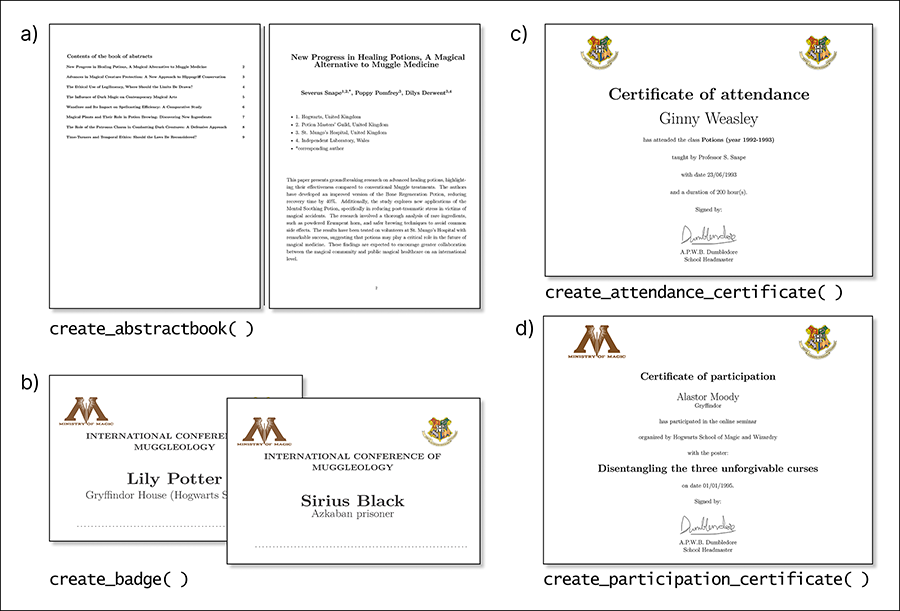
Figure 3.
Examples of outcomes from
each event-related function in labeleR. a) Abstract book: this function creates
a document with pages with the title, authors, affiliations and abstract
(variable fields) and can include a table of contents and front page. b) Badges:
they include the name and affiliation of each participant, a fixed field for
the title, the option to add two images on top, and a dashed line at the bottom
for additional hand-written information. c) Attendance certificate: attendee
name is a variable field, while event name, signer, and date are fixed fields.
d) Participation certificate: includes the participant name, affiliation and
title of the communication, plus several fixed fields. Both certificate
functions allow two images on top, a signature at the bottom, and offer Spanish
and English templates.
Figura 3. Ejemplos de los resultados de cada
función relacionada con eventos en labeleR. a) Libro de resúmenes: esta función
crea un documento con páginas que incluyen el título, los autores, las
afiliaciones y el resumen (campos variables), y además se puede incluir un
índice y una portada. b) Credenciales: éstas incluyen el nombre y la afiliación
de cada asistente, un campo fijo para el título, la opción de añadir dos
imágenes en la parte superior y una línea discontinua en la parte inferior para
información adicional que se escriba a mano. c) Certificado de asistencia:
incluye el nombre del asistente como campo variable, mientras que el nombre del
evento, la firma y la fecha son campos fijos. d) Certificado de participación:
incluye el nombre del participante, su afiliación y el título de la
comunicación, además de varios campos fijos. Ambas funciones de certificado
permiten dos imágenes en la parte superior, una firma en la parte inferior y
ofrecen plantillas en español e inglés.
Further applications
The labeleR
philosophy is quite simple: creating multiple documents with a common design
from a dataset containing the required information. It offers a modular
structure that allows for customization and extension for new applications. For
instance, the newly added create_multichoice( )
function generates multichoice tests randomizing the order of questions and
possible answers from a table including a set of questions. New developments
will happen in the GitHub repository (https://github.com/EcologyR/labeleR) and eventually pushed to CRAN.
Authors' Contribution
All authors have collaborated through the
process of conceptualization, software development and manuscript writing,
reviewing and editing. F.R.S. was responsible for founding during the
development of the main steps of the software.
Financing, required permits, potential
conflicts of interest and acknowledgments
This package has been developed
collaboratively between Sevilla and Madrid, with continuous feedback from
colleagues in both locations. We acknowledge their input, support and
collaboration. We are especially grateful to Manuel Molina for his ideas at labeleR initial stages. We would like to emphasize that the original idea
has been built horizontally among early career researchers. Our work has been
possible thanks to the support of the institutions and projects that employ us,
and especially by the software-developing workshop (Cádiz, La Muela, 2023)
organized by AEET and funded by US-1381388 grant from Universidad de
Sevilla/Junta de Andalucía/FEDER-UE.
J.G.A. was supported by Next Generation EU
Investigo contract (URJC-AI-17) and ANTENNA Biodiversa+ and European Commission
(PCI2023-146022-2) postdoctoral contract. J.M.M. was supported by the Comunidad
de Madrid and Universidad Autónoma de Madrid doctoral grant
PEJ-2020-AI/AMB-17551 and research assistant contract PIPF-2022/ECO-24251.
F.R.S. was supported by VI PPIT-US and grants US-1381388 from Universidad de
Sevilla/Junta de Andalucía/FEDER-UE and CNS2022-135839 funded by
MICIU/AEI/10.13039/501100011033 and by European Union NextGenerationEU/PRTR.
I.R.G. was supported by a doctoral grant at Universidad Autónoma de Madrid and
a postdoctoral position at Universidad de Sevilla (CNS2022-135839).
The authors declare no conflicts of
interest.
This note has been reviewed through an open
and collaborative process available at: https://github.com/ecoinfAEET/Notas_Ecosistemas/issues/60. We would like to thank David García-Callejas and Verónica
Cruz-Alonso for their helpful comments and suggestions.
References
Allaire, J., Xie, Y., Dervieux, C., McPherson, J., Luraschi, J., Ushey, K.,
Atkins, A., et al. 2024. Rmarkdown: Dynamic documents for r. Retrieved
from https://github.com/rstudio/rmarkdown
Banasiak, G., Charchut, B., Gould, M., Kaliszewski, B., Bialek, Q.M. 2022.
EntomoLabels v8.3.01. Retrieved from https://labels.entomo.pl/
Lafferty, D. 2025. LichenLabler v1.9.0.0. Retrieved from https://help.lichenportal.org/index.php/en/resources/printing-labels/
Pando, F., Lujano, C., Cezón, K. 2019.
Elysia: Programa de gestión de colecciones de biodiversidad (v.2.0).
Digital.CSIC. doi:10.20350/DIGITALCSIC/14520
Zhang, J., Zhu, H., Liu, J., Fischer,
G.A. 2016. Principles behind designing herbarium specimen
labels and the R package ’herblabel’. Biodiversity Science 24(12),
1345–1352.
![]() , Jimena Mateo-Martín3
, Jimena Mateo-Martín3 ![]() , Francisco Rodríguez-Sánchez4
, Francisco Rodríguez-Sánchez4 ![]() , Ignacio Ramos-Gutiérrez3,4,*
, Ignacio Ramos-Gutiérrez3,4,* ![]()